Fast SVD
Singular value decomposition is an expensive operation. For rectangular matrices with significant different dimensions, i.e. very “fat” or “thin” matrices, there is a trick to make the computation cheaper. This trick is implemented in fast.svd() of the R package corpcor.
Calculate SVD
The singular value decomposition of a matrix \(M\) of size \(m \times n\).
\[ M = UDV^T \]
\[ \begin{align} MM^T &= (UDV^T)(UDV^T)^T \\ &= (UDV^T)V(UD)^T \\ &= UD (V^TV) (UD)^T \\ &= UD(UD)^T \quad (V\text{ is orthogonal}) \\ &= UDD^TU^T \\ \end{align} \] Thus the decomposition of \(MM^T\) gives \(U\) and \(D^2\). The positive singular values of \(M\) can be inferred from \(D^2\). For this reason, fast SVD can only return positive singular values.
In the case of fat matrix \(M\) (\(n > m\)), \(MM^T\) is of dimension \(m\times m\), which is smaller than the original \(M\).
V is then calculated by
\[ V = M^T U D^{-1} \]
How fast can it be?
dat = data.frame('m' = rep(50, 6),
'factor' = c(1,2,5,10,20,50),
'svd' = rep(0, 6),
'fast.svd' = rep(0, 6))
dat[,'n'] = dat[,'factor'] * dat[,'m']
dat <- lapply(1:nrow(dat), function(i) {
x = rnorm(dat[i, 'm'] * dat[i, 'n']) %>%
matrix(nrow = dat[i, 'm'], ncol = dat[i, 'n'])
slow <- function() { return(svd(x)) }
fast <- function() { return(fast.svd(x))}
t0 = proc.time()
tmp <- svd(x)
t1 = proc.time()
tmp <- fast.svd(x)
t2 = proc.time()
dat[i,'svd'] <- (t1 - t0)['elapsed']
dat[i,'fast.svd'] <- (t2 - t1)['elapsed']
return(dat[i,])
}) %>%
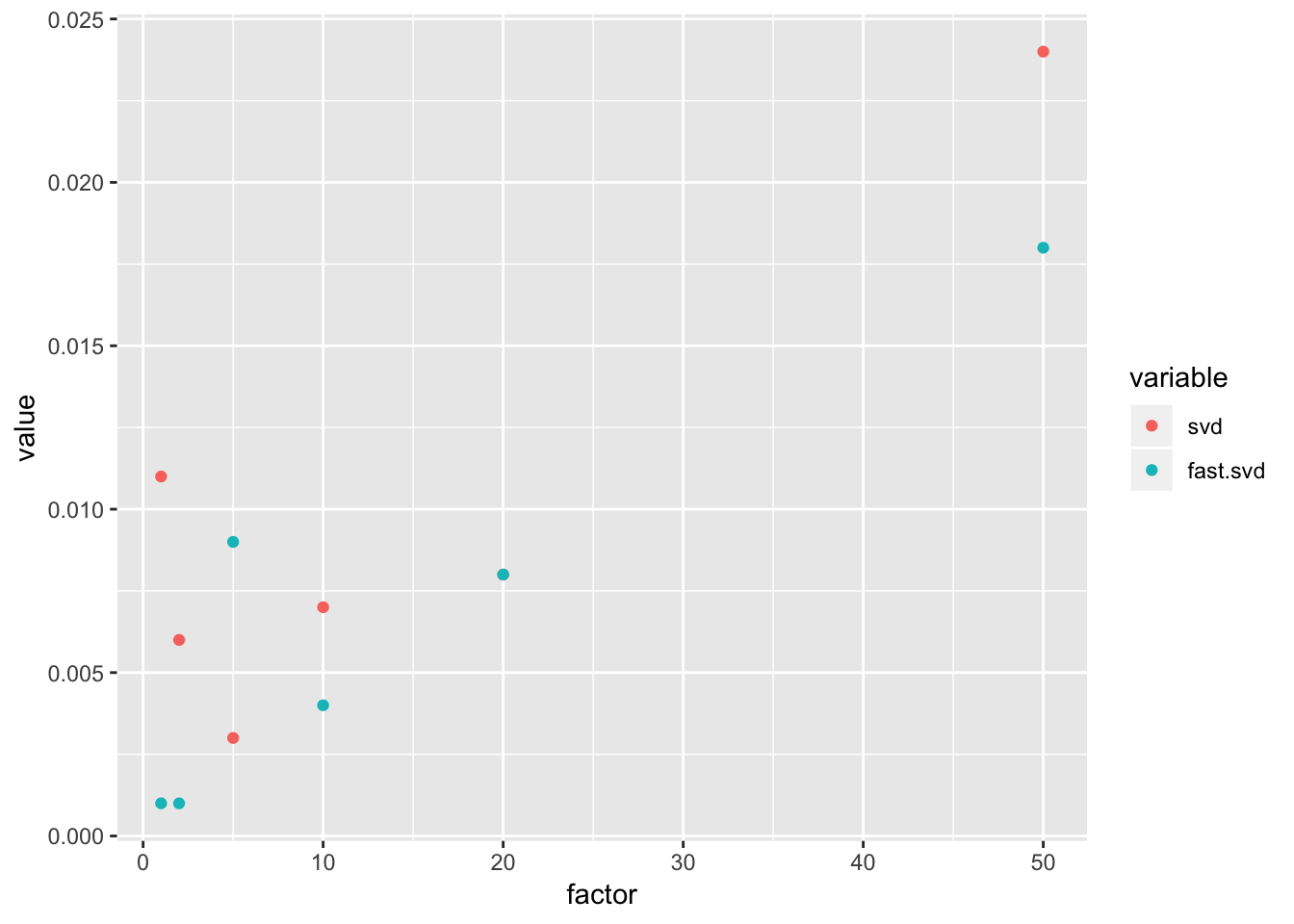
do.call(rbind,.)dat## m factor svd fast.svd n
## 1 50 1 0.011 0.001 50
## 2 50 2 0.006 0.001 100
## 3 50 5 0.003 0.009 250
## 4 50 10 0.007 0.004 500
## 5 50 20 0.008 0.008 1000
## 6 50 50 0.024 0.018 2500dat %>%
reshape2::melt(id.vars = c('m', 'n', 'factor')) %>%
ggplot() +
geom_point(aes_string(x = 'factor', y = 'value', color = 'variable'))