Fast Pearson Correlation
This note compares the performance of 2 methods for calculating Pearson correlation:
1. R stats::cor function
2. WGCNA::cor function (or corFast)
SparkR (1.6) provides a function corr to calculate the Pearson correlation between two columns of a data frame, but not between every pair of columns in a data frame. We would need to use Scala/Python interface for that.
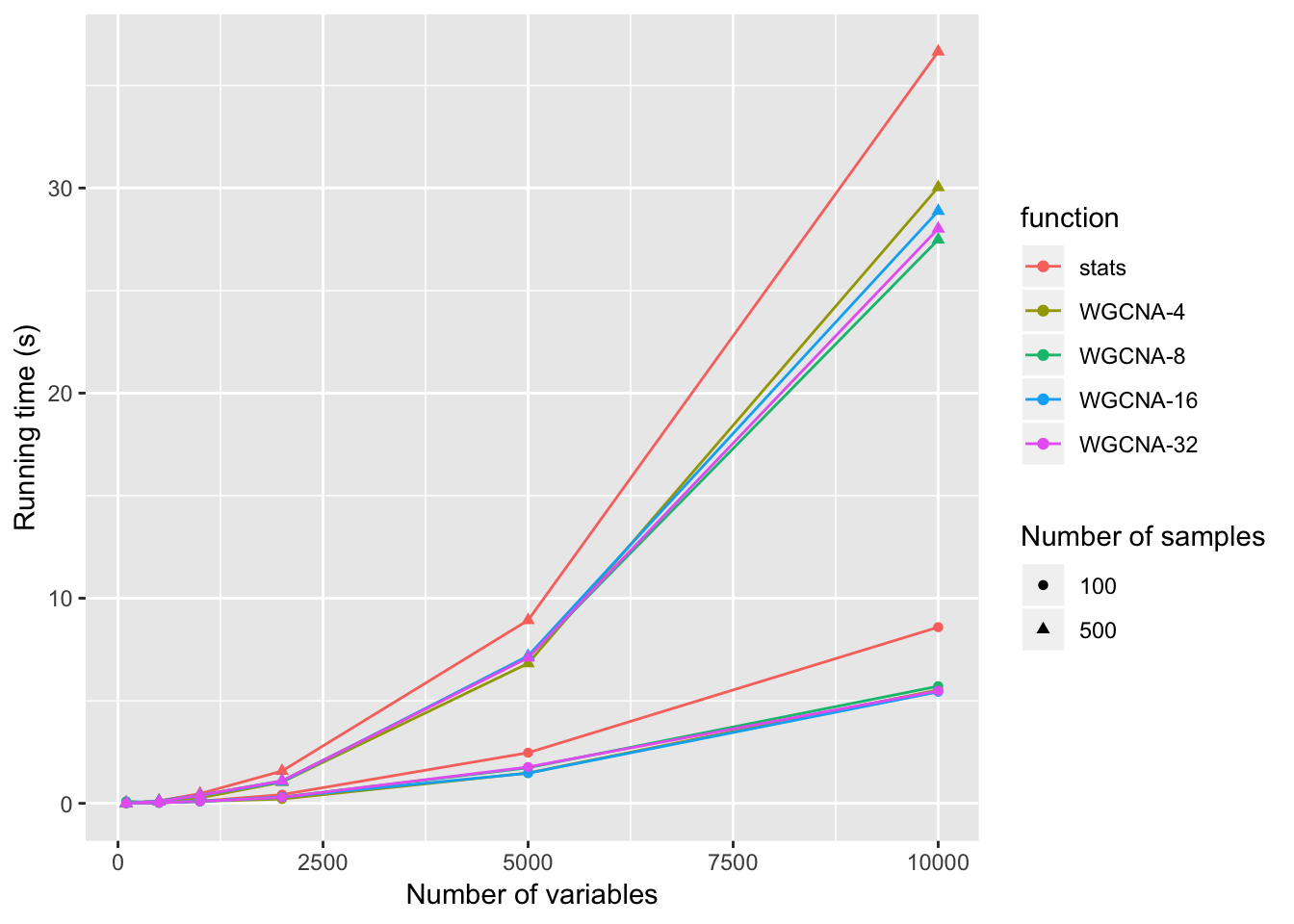
Correlation at 100 data points
library(WGCNA)
enableWGCNAThreads(nThreads = 32)## Allowing parallel execution with up to 32 working processes.m = matrix(rnorm(5000000),nrow=500)
nsamples = 100
times1 = data.frame(list('nvars' = c(100,500, 1000, 2000, 5000, 10000),
'nsamples' = rep(nsamples, 6)))
times1$stats = rep(0, length(times1$nvars))
for (i in 1:length(times1$nvars) ) {
nvars = times1$nvars[i]
times1[i,'stats'] = system.time(stats::cor(m[1:nsamples,1:nvars]))['elapsed']
for (nthreads in c(4,8, 16, 32)) {
times1[i,paste('WGCNA',nthreads,sep='-')] = system.time(WGCNA::cor(m[1:nsamples,1:nvars],nThreads = nthreads))['elapsed']
}
}Correlation at 500 data points
nsamples = 500
times2 = data.frame(list('nvars' = c(100,500, 1000,2000, 5000, 10000),
'nsamples' = rep(nsamples,6)))
for (i in 1:length(times2$nvars) ) {
nvars = times2$nvars[i]
times2[i,'stats'] = system.time(stats::cor(m[,1:nvars]))['elapsed']
for (nthreads in c(4,8, 16, 32)) {
times2[i,paste('WGCNA',nthreads,sep='-')] = system.time(WGCNA::cor(m[,1:nvars],nThreads = nthreads))['elapsed']
}
}
times = rbind(times1,times2)Timing result